
Bone marrow failure NGS panel
Download specifications as pdf
Targeted sequencing with the Ion Torrent System is able to identify single nucleotide variants, small insertions and small deletions. Variants in repeat sequences, large homopolymers and large insertions/deletions are not or difficult to identify.
The designed BMF Panel (IAD156379_243) consists of 3140 amplicons and is covering 861,9 Kbase. 99,1% of desired areas (exons, flanking intronic regions, untranslated regions and promotor areas) are covered from the following 179 genes*:
Overview genes present in BMF-panel |
|||||||||
ABCAl |
BLOC1S3 |
CXCR4 |
FANCE |
HPS1 |
MAP2K2 |
PMS2 |
RPL35A |
SAMD9L |
THP0 |
ABCB7 |
BLOC1S6 |
CYCS |
FANCF |
HPS3 |
MASTL |
PRF1 |
RPL36 |
SBDS |
TINF2 |
ABCG5 |
BRCA1 |
DDX41 |
FANCG |
HPS4 |
MKL1 |
PTPN11 |
RPL5 |
SBF2 |
TP53 |
ABCG8 |
BRCA2 |
DHFR |
FANCI |
HPS5 |
MLPH |
RAB27A |
RPS10 |
SEC23B |
TPM4 |
ACBD5 |
BRIP1 |
DKC1 |
FANCL |
HPS6 |
MPL |
RAC2 |
RPS14 |
SH2D1A |
TPP1 |
ACD |
C15orf41 |
DNAJC21 |
FANCM |
IKZF1 |
MTHFD1 |
RAD51 |
RPS15 |
SLC37A4 |
UBE2T |
ACTB |
C6orf25 |
DTNBP1 |
FAS |
IL2RG |
MYO5A |
RAD51C |
RPS15A |
SLFN14 |
UNC13D |
ADA |
CBL |
ELANE |
FASLG |
ITK |
NBEAL2 |
RBM8A |
RPS17 |
SLX4 |
USB1 |
|
CDAN1 |
EPCAM |
FlI2 |
JAGN1 |
NBN |
RIT1 |
RPS19 |
SMARCD2 |
VIPAS39 |
AK2 |
CDC42 |
ERCC4 |
FYB |
JAK2 |
NF1 |
RMRP |
RPS24 |
SOS1 |
VPS33B |
ALAS2 |
CEBPA |
ERCC6l2 |
G6PC3 |
KLF1 |
NOLA2 |
RNF168 |
RPS26 |
SRC |
VPS45 |
ANKRD26 |
CEBPE |
ETV6 |
GATA1 |
KRAS |
NOLA3 |
RPL10 |
RPS27 |
SRP72 |
WAS |
AP3B1 |
CLPB |
EVl1 |
GATA2 |
LAMTOR2 |
NRAS |
RPL11 |
RPS28 |
STX11 |
WDR1 |
AP3D1 |
COX4-1 |
FADD |
GFl1 |
LIG4 |
PALB2 |
RPL15 |
RPS29 |
STXBP2 |
WIPF1 |
ASXL1 |
CSF2RA |
FANCA |
GFl1B |
LYST |
PARN |
RPL26 |
RPS7 |
TAZ |
WRAP53 |
ATM |
CSF3R |
FANCB |
GINS1 |
MAD2L2 |
PGM3 |
RPL27 |
RTEL1 |
TCIRG1 |
XIAP |
ATRX |
CTCl |
FANCC |
HAX1 |
MAGT1 |
PLAU |
RPL27A |
RUNX1 |
TERC |
XRCC2 |
BLM |
CTSC |
FANCD2 |
HOXA11 |
MAP2K1 |
PLCB2 |
RPL31 |
SAMD9 |
TERT |
YARS2 |
Genes present in BMF-panel
The percentage of target bases that is covered at least 20 times (%Base20x) is at least 99,0% for the recommended Mapped Reads of 4.000.000. For 69 different genes a few bases are missed, either in the design or due to practical coverage, as listed in Part 1 of the table below. Some pathogenic variants listed in the HGMD database are missed as well, as can be seen in part 2 of this table.
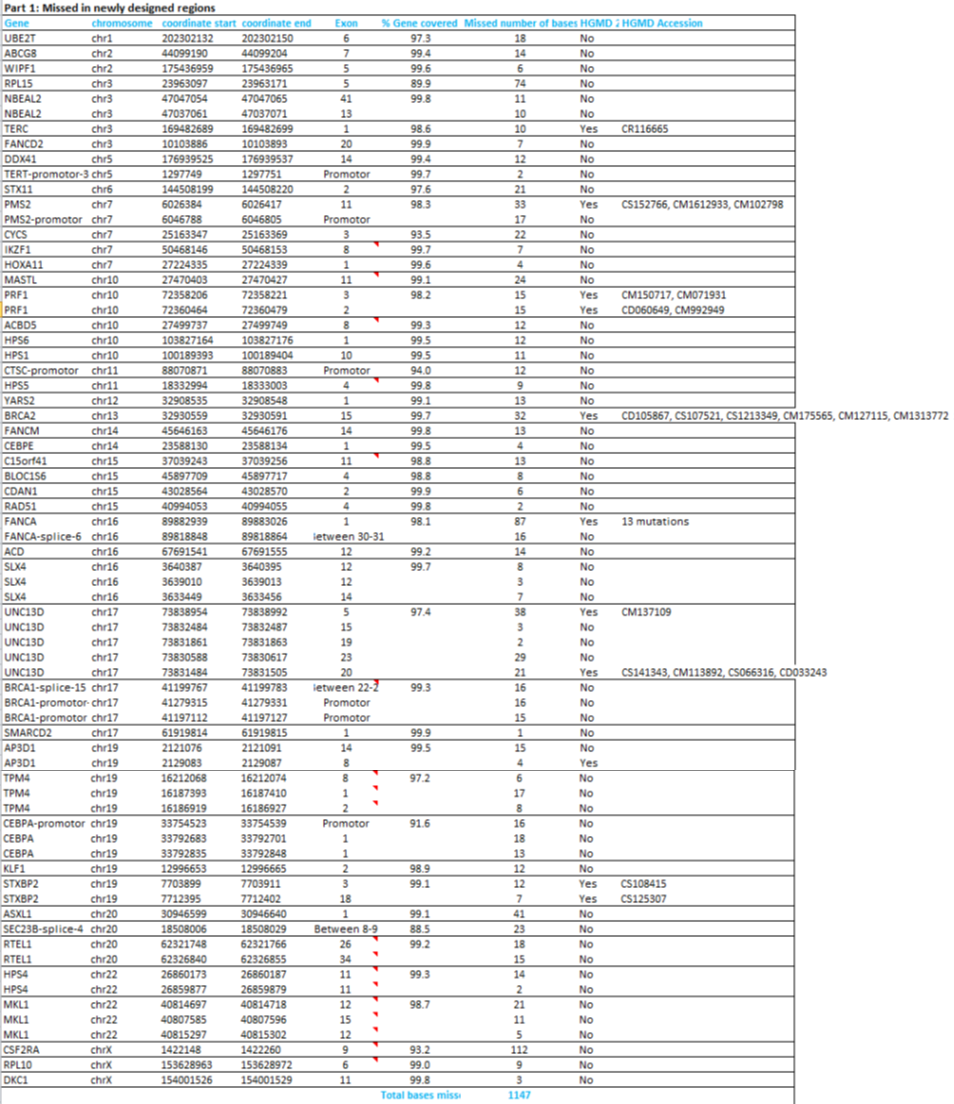
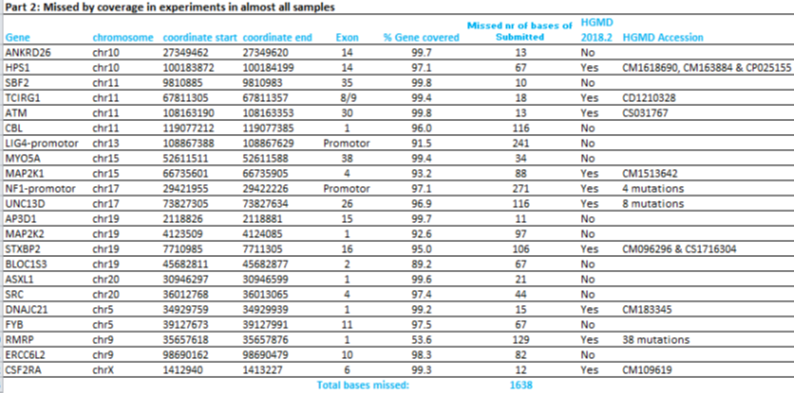
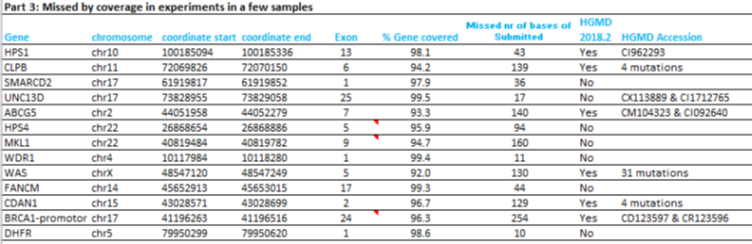
Some additional regions might be missed in a sample, due to low coverage or uniformity (part 3, below).
* 20 June 2025: Contrary to what was previously stated on this website, the ADA2 gene is not and has not been part of the bone marrow failure panel. For Sanger sequence analysis of the ADA2 gene, please contact the dept of clinical genetics at the AmsterdamUMC ([email protected], +31 (0)20 - 566 5281).
Background information
Service pages NGS

